The Molecular Virology Research Group of the Department of Medicine and Life Sciences, Pompeu Fabra University (MELIS-UPF), led by Juana Díez, and the Epitranscriptome and RNA dynamics group of the Center for Genomic Regulation (CRG), led by Eva M. Novoa, have led a study on how viruses reprogram the host cellular machinery to their advantage in order to reproduce more efficiently.
Specifically, the scientific teams have observed that viral infection induces a reprogramming of cellular tRNA modifications to favor the translation of viral codons. Thus, they maximize the expression of their proteins and, therefore, viral infection.
The paradox of efficient viral translation
Viruses are completely dependent on the host cell’s translation machinery. To express viral proteins from their genetic material, viruses need the cell’s tRNAs (transfer RNA). These detect the codons of the viral RNA that determine one or another amino acid.
The genetic code that establishes the correspondence between each amino acid and each codon is redundant, allowing an amino acid to be encoded by more than one codon. The frequency of these synonymous codons is not random; instead, it is biased and organism-specific. As the concentration of tRNAs in each organism is adapted to this frequency, sub-optimal codons slow down translation because the corresponding tRNAs are low in abundance compared to the tRNAs that recognize the optimal codons.
Viral dependence on cellular tRNAs is especially critical for positive-stranded RNA (RNA+) viruses whose first step in the infection cycle is precisely the translation of viral RNA. The group of RNA+ viruses includes many human pathogenic viruses, including new and emerging viruses such as SARS CoV-2 or the mosquito-borne viruses of dengue, zika or chikungunya.
All of these viruses express their proteins at very high levels. However, the genomes of these viruses are enriched in suboptimal codons and should therefore be translated inefficiently. To resolve this paradox, the authors used the chikungunya virus as a study model because of its high level of replication.
The secret lies in the epitranscriptome
The MELIS-UPF and CRG work, which also involved the collaboration of researchers from the University of Tartu, found that infection by the chikungunya virus induces cellular stress that leads to increased production of the tRNA modification enzyme KIAA1456. This leads to changes in the epitranscriptome (the set of chemical modifications that RNAs undergo) that favor the recognition by cellular tRNAs of suboptimal codons, present in cellular stress response proteins and also in the virus. This way, these codons become optimal. Therefore, the chikungunya virus genome has evolved so that the composition of its codons becomes optimal under the conditions of host cell infection.
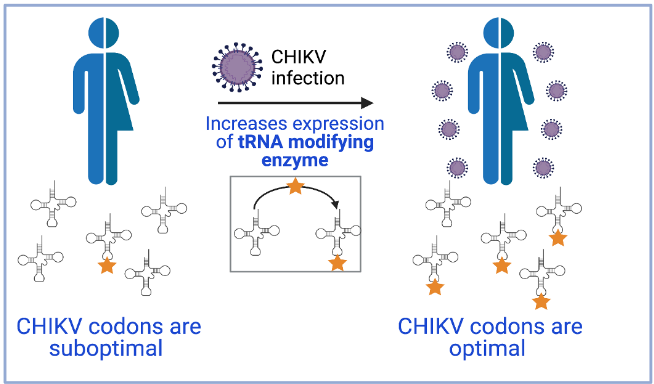
“Although the study has focused on the chikungunya virus, we have confirmed that other viruses use a similar strategy to favor the expression of their proteins, so we propose that the modification of tRNAs induced by viral infection is a general mechanism followed by many viruses”
Juana Díez, head of the MELIS-UPF group and principal investigator of the study
The experiments have been performed with established cell lines of human liver cells, embryonic kidney cells and kidney fibroblasts. The discovery puts tRNA modification enzymes in the spotlight for the study of other viruses and new therapeutic targets for the development of broad-spectrum antivirals.
Jungfleisch J et al. CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nature Communications. August, 2022. DOI: 10.1038/s41467-022-31835-x.