Reproducibility is basic for science, since new research is based on previous knowledge, which must be robust – and therefore, reproducible. In computational biology, it is essential to be able to understand the code written in order to reproduce an analysis. However, the reasoning behind the code is sometimes given for granted and not enough attention is paid to make clear how it was created.
“Non reproducible single occurrences are of no significance to science”
Karl Popper
The Centre for Genomic Regulation (CRG) organised a workshop last April in this context. Very practical in nature, the workshop brought together 30 people – PhD students, bioinformaticians and undergraduates, mostly MSc students – who wanted to learn how to make their research more reproducible, transparent and collaborative.
Ulf Tolch, from QUEST Centre in Berlin, Germany, was in charge of giving this 4h interactive seminar, that took place on April 9 in the Charles Darwin room of the Barcelona Biomedical Research Park (PRBB). Participants were mostly coming from centres at the PRBB – specially from the CRG and CNAG, with some from Hospital del Mar Medical Research Institute (IMIM) and Institute for Evolutionary Biology (IBE: CSIC-UPF) – although some were from external centres (UB, BSC, IBEC).

After an initial introductory round where everyone explained what type of data they work on, Tolch asked them how confident they were about the reproducibility of their research. The answers were clear evidence of the need of this course.
He started off with a general view on the importance of data management, discussing with the participants about the very important “Data management plans”. Most agreed that these data management plans are very useful – and compulsory for many funders – but in some cases they may be difficult to write, for example in cases of exploratory research, where you do not know what you will find.
The course instructor showed the audience several data repositories and related resources that they could use to make their data open:
- Re3data.org – a registry of data repositories to find the most appropriate one for your data
- OSF (Open Science Framework) – more of a project manager than a data repository, it is good for pre-registration of projects and to share the research process in general
- Zenodo – housed at CERN. It allows to store snapshots of your github repository, which is useful because it changes with time, and you may want to have a view of how it was when you published your paper, for example.
- Dryad – a general purpose repository
- Figshare (company owned)
- GitHub (on the cloud)
- GitLab (on your server)
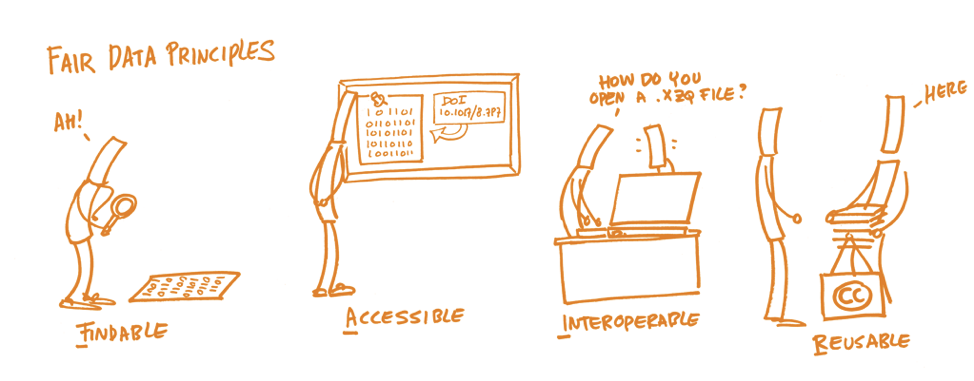
Making your data FAIR
These open data repositories help ensuring your data follows the FAIR principles, which Tolch emphasized were very important to make research reproducible:
- Findable
- Accessible (both for humans and machines)
- Interoperable (both for humans and machines)
- Reusable – add a license so people know they can reuse it!
Once the first theoretical part was over, the participants started working, each on their laptop. After pointing out several issues to think about before starting a collaborative project – such as thinking beforehand what your audience will need to reproduce your work – Tolch showed them how to use GitHub, a platform that allows users to store their projects using the control version system of Git.
The participants learned in a friendly and relaxed atmosphere how to invite collaborators, how to create branches of their code – to try things out, or to divide the work into smaller parts and give each part to a collaborator – and then how to merge them back to their master branch. Importantly, Tolch reminded them to write a commit message everytime they commit (that is, everytime they made a change to the code), so that anyone (including their future self!) knew what had been changed.
How to write a paper
The last part of the seminar touched on how to merge all the data together in order to write a scientific paper that has all the necessary information for others to reproduce the study. For this, Docker containers help. As Tolch asked; what happens if someone downloads our code in 5 years and the software version has changed, the library from where we got the data has changed slightly, the name has changed…? The figures of the paper will not be reproducible. The way Docker container solves this is by packaging your data together with the specific library version, R version, linux version, etc. used for the study. Like this, you or others will be able to ‘reproduce’ later on the exact same environment you have now.
For the final part of the seminar, the audience divided into three groups to have a quick look at three platforms that simplify usage and put together technologies such as Docker containers or Jupyter notebooks:
The last minutes were devoted to saying three things they had learned and three things they hadn’t learned that they wished they had. One of the participants summarised it very well: “I learned there’s a lot to be learned!”.
All in all, the students said they wanted more of this type of workshops – including the opportunity to bring their own data and work with them, but that the seminar “gives a good overview of things that you need to learn if you want to be able to make your research clean and reproducible”. Indeed, the participants left the seminar room at the PRBB with a lot more confidence on how to ensure the robustness of their research.
This workshop took place thanks to the Foster H2020 project in which the CRG is a partner. On the project webpage you can find many more training resources regarding open science.
Check out this video for an interview with the trainer, Ulf Tolch.