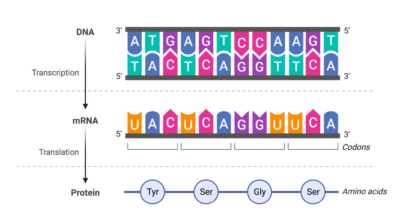
In a recent study, researchers from the Centre for Genomic Regulation (CRG) developed a new method to measure RNA modifications in cancer, which could lead to new approaches in the detection of this disease.
We talked with the first authors of the study, Oguzhan Begik and Morghan Lucas, from Eva Novoa‘s group, in order to know more about this promising method.
Why are RNA modifications important in the study of cancer?
RNA modifications regulate many aspects of RNA biology; structure, degradation, localisation, translation, splicing,… the list goes on. These chemical alterations have recently been shown to play key roles in many biological processes, such as development, immunity and cancer progression.
“There are over 170 RNA modifications identified thus far, which decorate all types of RNAs in a dynamic fashion”
Oguzhan Begik, CRG, co-author of the study
In a previous study from our lab we found that enzymes responsible for placing, removing and recognising RNA modifications are dysregulated in multiple cancer types and may provide novel cancer biomarkers or therapeutic targets.
Indeed, recent work provided proof-of-concept that the targeting of RNA modifying enzymes presents a promising new avenue for anti-cancer therapy in acute myeloid leukemia, and we anticipate this research space will gain a lot of traction in the coming years with the advancement in third-generation sequencing technologies.
How could these findings lead to new approaches in cancer detection?
The first step of characterising the functional role of RNA modifications in various biological processes is detection. We need to understand which RNA modifications play a role in cancer progression and metastasis, and when. Then we can then use this knowledge to identify potential biomarkers to detect cancer or to better stratify patients.
“By using RNA modification profiles we could inform cancer prognostics and exploit dysregulated RNA modification enzymes as therapeutic targets.”
Morghan Lucas, CRG, co-author of the study
Could your new method be useful in other fields of research and diseases?
In our latest study, we expand upon our previous efforts of detecting RNA modifications in native RNA molecules in an unbiased transcriptome-wide manner, by expanding the repertoire of modifications we can detect, as well as quantifying their stoichiometry.
This method is not limited to specific samples or by disease type; it can be applied to a wide range of biological systems, from C. elegans to patient samples, to study RNA modification dynamics in native RNA molecules.
Begik, O., Lucas, M.C., Pryszcz, L.P. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol (2021).