This text was written by Ivan Milenkovic, a PhD student at the laboratory led by Eva Novoa at the Centre for Genomic Regulation (CRG). Ivan won the 1st award in the Rin4′ science outreach contest with his presentation “La vida dinàmica dels ribosomes” (“The dynamic life of ribosomes”). Here, he tells us more about his research into ribosomes, cellular organelles involved in the formation of proteins and that, until relatively recently, were thought to be static and uniform in all tissues. Ivan also shares his experience as a PhD student so far.
You can also read an interview with him here.
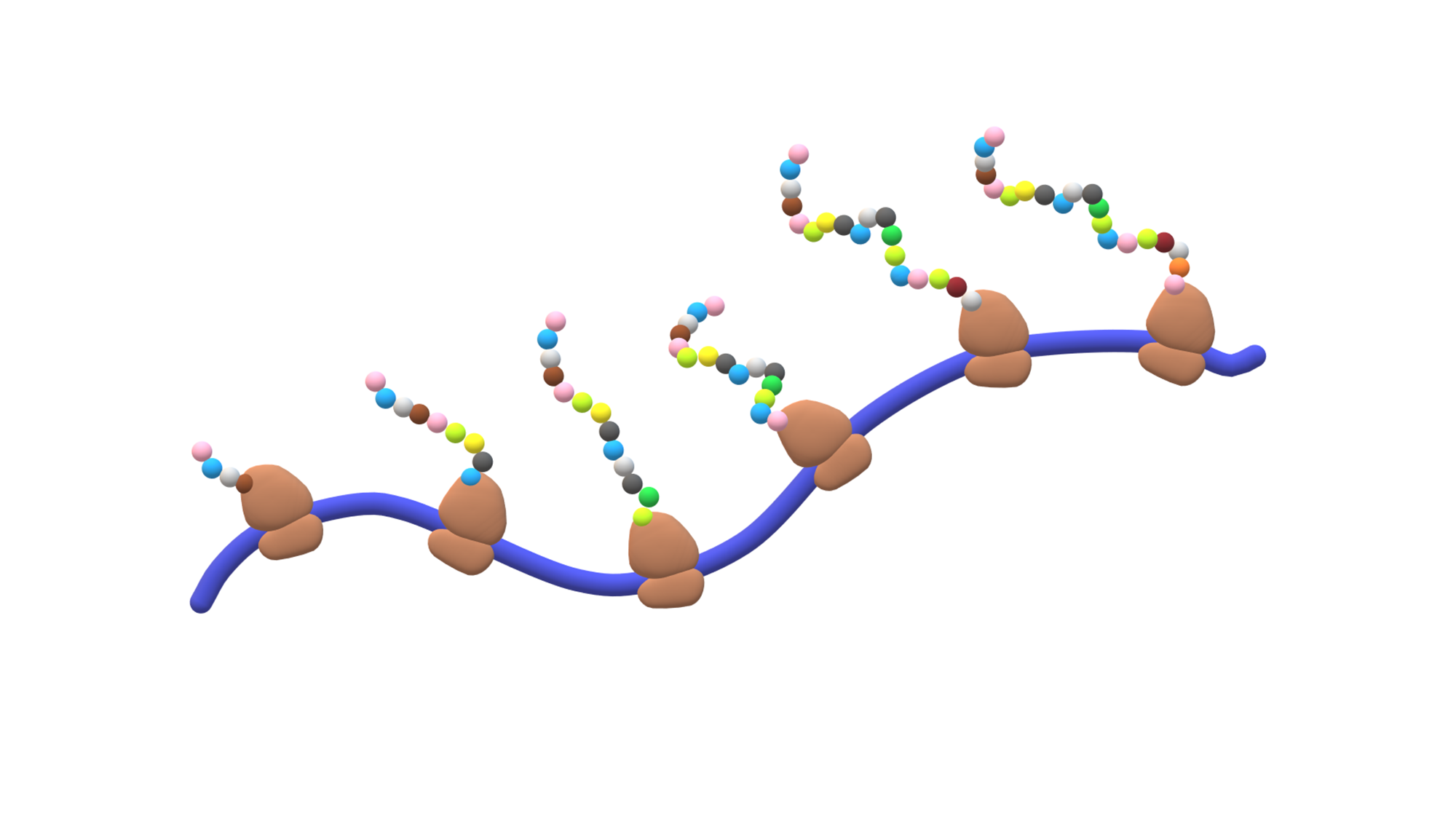
The aim of my project is understanding the biological role of ribosome heterogeneity. For a long time we thought that ribosomes – the molecular machines reponsible for protein synthesis in all living beings – tend to be uniform, with a static composition that is identical in all tissues in our bodies. However, recently it has been discovered that ribosome structure can be a lot more heterogeneous, and that this heterogeneity can be tissue- or developmental stage-specific. I am trying to understand whether this structural diversity infers a specialized function.
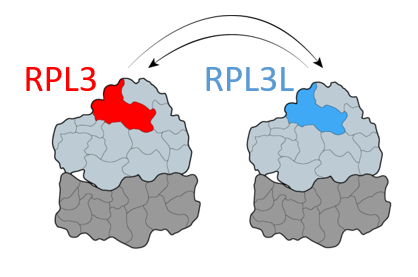
Some ribosomal proteins have paralogs – proteins stemming from one ancestral gene that was duplicated during evolution. We have found that a ribosomal protein called RPL3L is only expressed and incorporated into ribosomes in striated muscles, as opposed to its paralog, RPL3, which is expressed in all the other tissues. By generating a Rpl3l knockout mouse model (a mouse that does not have the Rpl3l gene), we have identified that in the absence of RPL3L, RPL3 is expressed in striated muscles and compensates for the loss of its paralog. Furthermore, we have found that the ribosome-mitochondria interactions change depending on which protein is incorporated into the ribosome, suggesting that the choice of ribosomal protein paralog can affect ribosome function.
Ribosome heterogeneity can also stem from rRNA, notably from rRNA modifications. In the lab, we have developed a method to study RNA modifications by sequencing native RNA molecules using the Oxford Nanopore Technology platform. We have sequenced the rRNA from several mouse tissues at different developmental stages and found that several nucleotides are modified in a dynamic manner, both across tissues and developmental stages.
Altogether, these results go in line with several studies from the recent years which depict ribosomes as a lot more dynamic than we previously thought.
Wet and dry lab, an exciting challenge
I am currently in the 4th year of my PhD. The main challenge so far was learning (to learn) bioinformatics while doing experiments at the same time. Having a background in biochemistry, I had almost no experience in bioinformatics when I started my PhD. However, my supervisor, Eva Novoa, highly encourages all PhD students to be involved in both the wetlab and the bioinformatics parts of the lab, so from my first days in the lab I was immersed in this environment that helped me work on both in parallel. This was also the most rewarding – I really enjoy taking part in all stages of the project: from the very first experimental designs to the very last bioinformatics analyses and visualization!