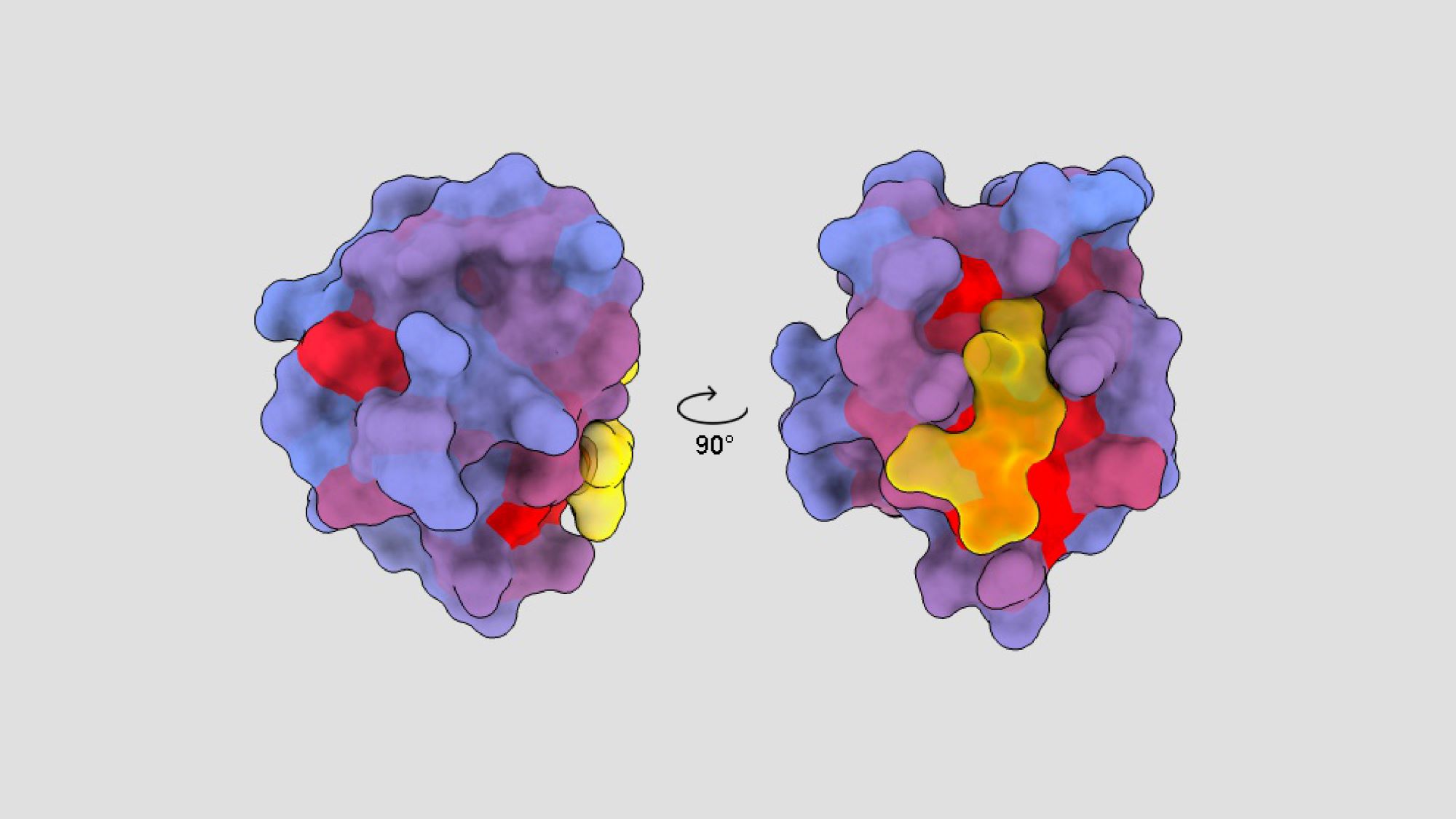
This image by André Faure, a postdoctoral researcher at the Centre for Genomic Regulation (CRG), shows the structure of a human protein, with its active sites marked in yellow and allosteric sites in red and blue, the latter ones discovered thanks to a new technique.
Most drugs are designed to bind to the active site of the target protein, inhibiting or activating it. The problem is that these active sites are usually very similar between different proteins, so the drug can be attacking several proteins at the same time, which produces side effects and reduces efficiency.
There are other places in proteins, more hidden but more specific, which, when bound to a drug, also modify the action of the protein. These are called allosteric sites, and they are normally almost impossible to detect.
Now a research team from the CRG has developed a new technology that makes it possible to discover these ‘hidden’ allosteric sites in the protein’s structure. The technique involves changing each and every one of the protein’s amino acids (the protein’s building blocks) one at a time, and seeing, for each of the thousands of resulting versions of the protein, what the effect is when introduced in living cells in the laboratory.
“It’s a ‘brute force’ experiment that allows us to find at least one ‘weak spot’ in each protein, a place we could potentially target with a drug. Even for those proteins that until now were considered ‘undruggable” explains Ben Lehner, leader of the group.
Furthermore, according to Júlia Domingo, first co-author of the article, this technique is “more efficient, cheaper and faster” than others currently used, and could revolutionize drug development.
The researchers have used the technique to study these places or ‘hidden pockets’ in two of the most common protein domains in humans, SH3 and PDZ. They have seen that these allosteric sites can not only be turned on and off, but also modified in more subtle ways, as the temperature control in a thermostat. This could help develop much more precise and intelligent drugs.
Want to see your photo here? Send us images related to science or life at PRBB a ellipse@prbb.org.
Faure, A.J., Domingo, J., Schmiedel, J.M. et al. Mapping the energetic and allosteric landscapes of protein binding domains. Nature 604, 175–183 (2022). https://doi.org/10.1038/s41586-022-04586-4