Is it possible to know the identity of a cell, to learn how it works, by knowing which genes are expressed in it? This would be the aim of the Biodiversity Cell Atlas project, but not just for a cell but for all the different cell types present in living beings.
This project, conceived following an expert meeting held at the Centre for Genomic Regulation (CRG) in May 2023, proposes to map all the cell types of different species; i.e. an atlas of cellular biodiversity that will provide knowledge in terms of evolution, industrial applications, advanced therapies, synthetic biological systems, conservation strategies or simply a better understanding of how cells function.
The approach is as ambitious as the Human Genome Project or the Human Cell Atlas. By bringing together all our knowledge of cells, we will be able to better understand the diversity and evolution of life.
However, the Cell Biodiversity Atlas faces several challenges:
- on the one hand, how to unify or standardise the methods used to compare different cell types, as there is no single method that works for all cells in all organisms;
- on the other hand, how to process and store the whole set of data obtained, and how to make it accessible to the entire scientific community.
That’s why the research group led by Arnau Sebé-Pedrós at the CRG, based at the Barcelona Biomedical Research Park (PRBB), has received a 3.8 million dollar grant from the Gordon & Betty Moore Foundation to carry out the preliminary work to develop and implement the techniques to create the atlas. This will be done in collaboration with teams in Oxford, Norwich, Chicago and Heidelberg (Germany).
We spoke to Sebé-Pedrós about the challenges involved and the importance of this major project, for which collaboration between the scientific community will be crucial.
What is the main objective of the Biodiversity Cell Atlas and why did you decide to undertake this project?
Well, actually this atlas is just a proposal at the moment, there is no funding for it yet… But the idea came out of a meeting I organised here at the PRBB last year, supported by the Welcome Trust, where a lot of us working on single cells in non-model organisms met. We started discussing that it would be a good idea to create this atlas because we are a growing community – at the moment there are maybe 20 to 40 groups around the world doing these transcriptomic cell atlases – and up until now everyone has been doing it in their own way. There were no standards so it’s difficult to compare different cases. We also wanted to stimulate discussion forums on methods, data interpretation, etc. and build a community.
Since then, we have continued to work with working groups where we meet every two months to put things together. And now we have this funding for the next 3 years to do a very specific task; to develop methods, both experimental and analytical, and to work with the EBI (European Bioinformatics Institute) so that it can eventually be the home of all the data.
So what you are doing is a pilot test?
You could put it that way, yes. This project should be the basis for the Cellular Atlas of Biodiversity, if we can show that it makes sense, that it is feasible, that it can be done with current methods… We need to demonstrate that we have an experimental and computational strategy and a database where we can store all the data.
What do you see as the main challenges?
There are several. Sample preparation is difficult. To cover the full range of biodiversity, samples need to be taken from different species in their natural habitat – in some cases this means on a ship at sea, and a sample that may only last a few hours. And sometimes there may not be enough sample material.
Then the transcripts expressed in each cell need to be sequenced– that is, all the genes that are present in the form of RNA in a cell at a given time and that give the cell its identity. And the computational analysis needs to be carried out to understand which cell each transcript belongs to.
But just as the challenges in genomics projects have been overcome (until recently we had dozens of sequenced genomes, now we have thousands!), I am sure they can be overcome in this project.
And what technologies will be needed for this project?
We are using single-cell transcriptomic sequencing – this technology has been around for about 10 years and is being used more and more. We believe that today it is already possible, in terms of cost and scalability, to use it for large projects like this one.
We will also be doing comparative transcriptomics, looking at how different species do cell transcription, which genes they express in different situations. It is a challenge, but I see it as an opportunity rather than a limitation. We have to start doing it, and we will see how we overcome any difficulties.
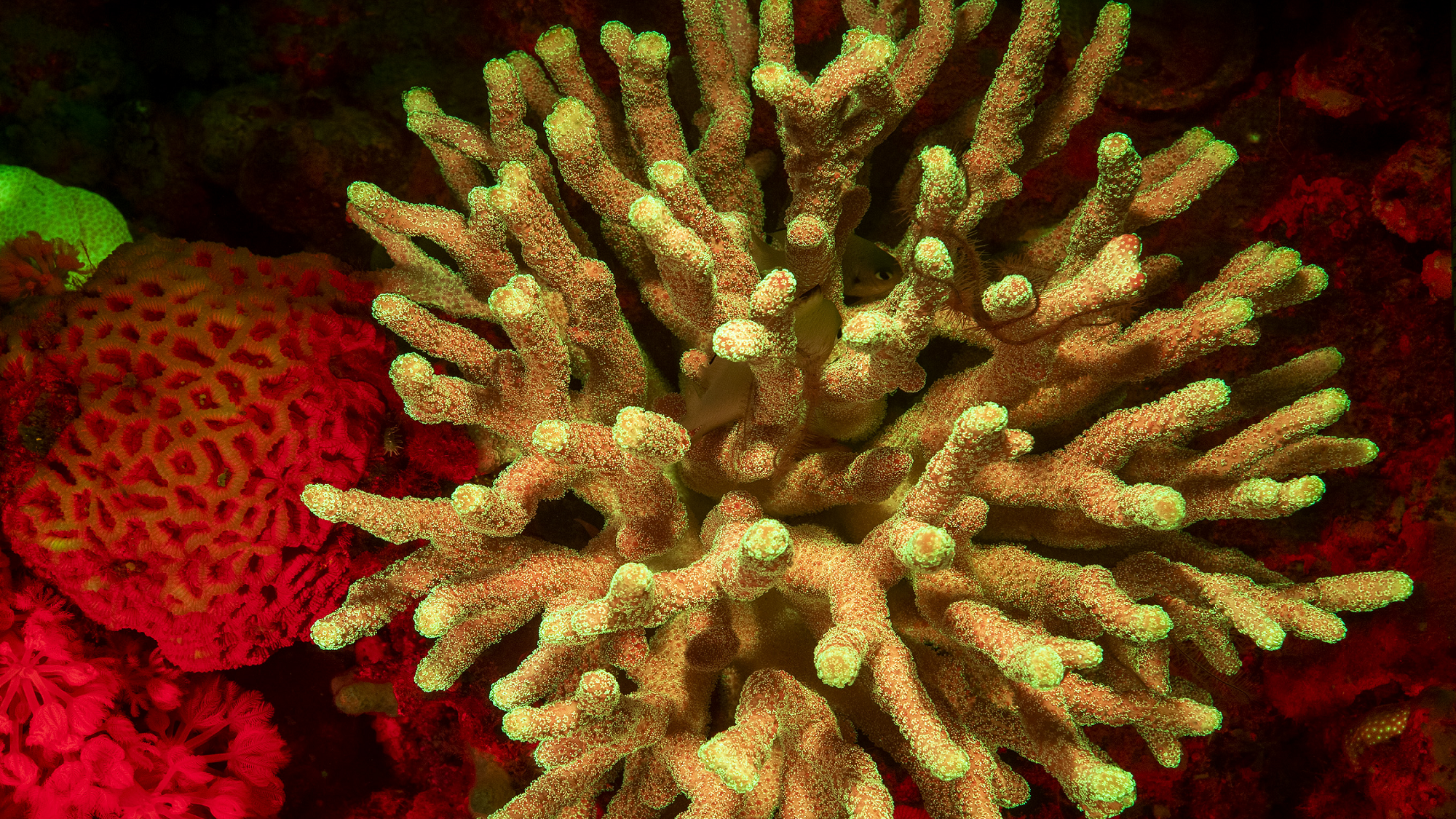
We don’t know what this data will allow us to do – we have an intuition, for example, that it can help us to understand how genes work, how they interact with each other (for example, if these two genes are always expressed together during evolution, they probably have an important function together). Also for conservation issues; for example, we are looking at how corals respond to thermal stress, how their transcription varies under these conditions, which can model climate change.
Single-cell transcriptomic sequencing has been around for about 10 years and we believe it is now possible to use it on a large scale. There are challenges, but I see them as opportunities rather than limitations.
Arnau Sebé Pedrós, CRG
Which organisms are you using for this first test?
We are going to start with organisms that we already know about, that we have data on, that we have protocols on, that represent different difficulties that might arise, to see how we can achieve this. These include well-known organisms such as the C. elegans worm or the Drosophila fly, but also a mollusc, an annelid, a sea urchin, a sea anemone, brown algae, a fungus and a liverwort (a type of herbaceous plant).
The requirements are that we have easy access to plentiful material and that we have a high-quality genome sequenced.
And what do you expect to achieve in the next few years?
With these ‘easier’ organisms, we will test and compare methods that can be useful for cell atlases in non-model organisms, in order to build a ‘decision tree’ that will help us approach new species in the future. This ‘tree’ will consist of a kind of guide to decide which steps to take with each species: i.e. if your chosen species has certain favourable conditions (in terms of availability, genome quality, etc.), we propose protocol X; if the conditions are not so good, another, more limited protocol; depending on the conditions, it may not even be necessary to try it… We will also propose quality controls, metrics to know if you are on the right track.
And above all, we hope to show that it is worthwhile to expand the project and carry out the Atlas of Cellular Biodiversity!
Article written in collaboration with Diana Darriba.