Jana Selent studied Pharmacy and in her PhD she specialized in drug design. Her curiosity to understand how drugs work at the molecular level and her desire to observe these interactions led her to become interested in bioinformatics. Currently, she leads the GPCR drug discovery group of the Biomedical Informatics Research Program (GRIB) of the Hospital del Mar Research Institute.
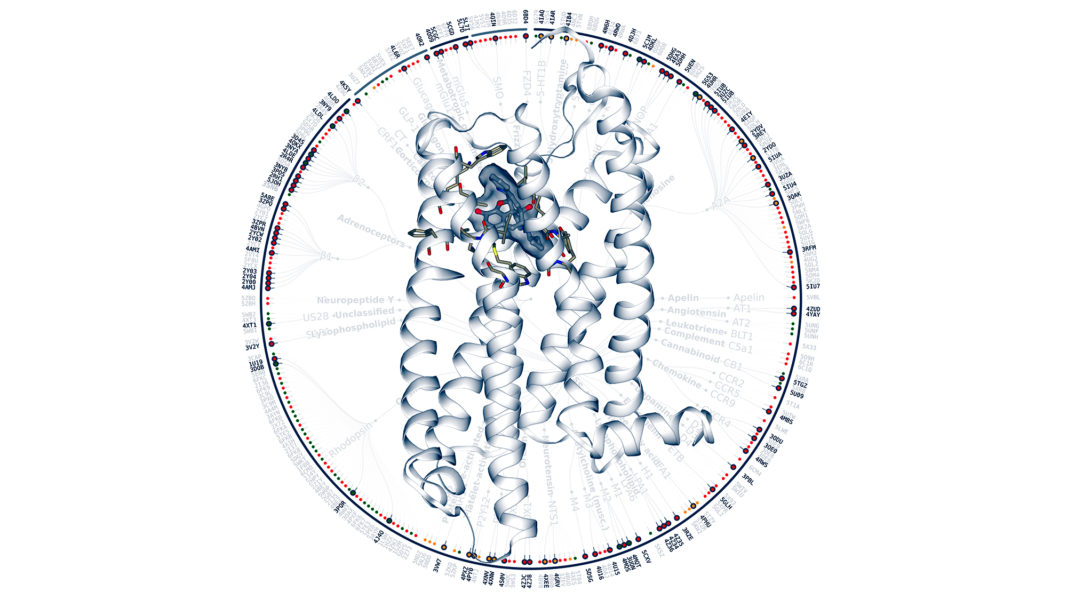
Her group is looking for drug targets on G-protein-coupled receptors (GPCRs). GPCRs are proteins found in the cell membrane that interact with substances outside the cell to produce changes in the cell. Therefore, they act as receivers of external signals. By studying how these outward signals produce changes, Selent’s group wants to help find drugs for mental disorders such as schizophrenia that are more specific and effective.
“I’ve been studying these receptors for 15 years and they haven’t bored me yet. Their workings are much more complex than we had imagined and we continue to make discoveries”
Jana Selent, GRIB
An abundant and key type of receptors
GPCRs are proteins that act as membrane receptors to transmit information from the outside to the cell. This information can come in the form of proteins, neurotransmitters, or even light or mechanical stress. Different external signals activate GPCRs, which in turn activate cascades of reactions that help cells adapt to external changes and survive.
There are about 800 different GPCRs and they are very important for the physiological processes that occur in our body. That is why they are very common targets for a wide variety of drugs, such as antihistamines or painkillers; and also other substances that generate changes in our body, such as caffeine. About 1 in 3 drugs target these proteins.
The function of many of these receptors is still unknown. “I’ve been studying these receptors for 15 years and they haven’t bored me yet. Their workings are much more complex than we had imagined and we continue to make discoveries”, explains Selent.
If we find out what their role is in the cell, we will have possible targets for new drugs. For this reason, much research focuses on these molecules. In addition, we can learn more about the interactions generated by the receptors we already know to improve existing drugs.
Observing how proteins interact
Proteins are the molecules that do most of the work in our cells and that is why they are the most common targets for drugs. For a drug to work, it must bind to its target in a specific way. This union is made because both substances fit together, like a key in a lock. Therefore, for drug development it is very important not only to understand what function proteins have, but also what their shape is and how interactions occur.
“There are experimental methods to reveal the 3D structure of a protein, but they give only a snapshot. We want to see how the structure of these proteins changes as they interact with other molecules”
Jana Selent, GRIB
Selent’s group uses bioinformatics to achieve dynamic visualizations of the shape of GPCR proteins when they interact with other molecules. “There are experimental methods to reveal the three-dimensional structure of a protein, such as crystallography or cryo-electron microscopy, but with them you only get a snapshot. We want to see the dynamics, how the structure of these proteins changes when they interact with other molecules”, explains Selent. Using bioinformatics, her group obtains three-dimensional models of proteins, which allow them to see how they move and how they interact.
Creating drugs and using artificial intelligence to screen candidates
The studies of Selent’s group, and that of several other international laboratories, have allowed the generation of a huge database of the structures of the GPCRs. With this database, screening can be carried out to see how different components, potential drugs that are designed virtually, interact with these receptors. The amount of information that is processed is so extensive and there are so many possibilities that artificial intelligence is needed to select the most relevant components.
Specialized companies synthesize these components, which the Selent Lab retrieved from a chemical space of billions of compounds . Then, they are tested in cell cultures by other research groups with which Selent’s group collaborates, such as teams from the Hospital del Mar Research Institute or other international centers.
Finding the right path
When an external signal activates a GPCR, it triggers a response in the cell by activating a cascade of interactions between molecules in what is called a signaling pathway. The drugs work by preventing or stimulating these responses. But the problem is that each receptor can activate different pathways.
The challenge is then to develop drugs that not only act specifically with the desired receptor, but also regulate only the desired pathway, without effecting other pathways of the same receptor. This helps avoid the side effects of the drugs. To do this, it is necessary to understand well what happens at the structural level in these molecules when they produce these responses.
The latest publication by Selent’s group helps advance the understanding of serotonin receptor 2A (5-HT2AR) signaling pathways. This receptor is a GPCR that can be used as a target for the treatment of schizophrenia. In their study, they have created receptor activating components to better understand its activation pathways.
They found that G⍺q protein activation is linked to memory loss while G⍺i1 activation is linked to psychotic behaviors. To arrive at this knowledge, they have combined computational methods with in vitro experiments, in cell cultures, and in vivo, using human brain tissue obtained postmortem. This greater understanding of receptor signaling pathways brings us closer to more effective schizophrenia treatments that minimize side effects.
Working collaboratively and ensuring transparency
Selent’s group works exclusively with computational models and collaborates with other groups that focus on the experimental part. Given the complexity of these signaling pathways, as well as the large number of receptors that exist, this collaborative work is essential.
“We want to encode the interaction of countless proteins that exist in the human body. This is a collective effort, something that could not be done by a single research group”, says Selent. She leads international projects and is one of the creators of the European Network for Research in Signal Transduction (ERNEST). This network consists of more than 500 members involved in GPCR research in the computational and experimental field.
“We want to encode the interaction of countless proteins that exist in the human body. This is a collective effort, something that could not be carried out by a single research group”
Jana Selent, GRIB
ERNEST members working in the computational field have created a large database with all the knowledge about GPCRs. The database contains dynamic models of the three-dimensional structures of the receptors and the components that can interact with them. This information can serve as a starting point for researching new drugs. The website can be visited to stream the dynamics of GPCRs in complex with these components.
The existence of this platform is also a way to promote good scientific practices, because it promotes the transparency and reproducibility of these data. The platform is used for research groups to show their results and shows the protocols used to obtain them. Selent is part of the PRBB’s Good Scientific Practices Group and has participated in the drafting of the protocol of good practices in the creation of simulations, which can be found in the methodology section of the platform.







