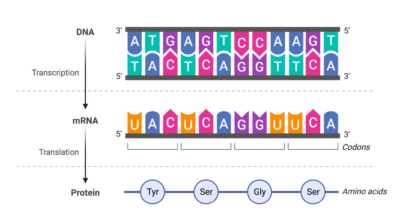
All living organisms produce proteins from their DNA, but not all of them are known. Some have an unknown structure or function, but the DNA sequence that encodes them is known. This sequence has been the basis of a collaboration between the Andalusian Center for Developmental Biology (CABD) and the Institute of Evolutionary Biology (IBE: CSIC-UPF) to analyze proteins using artificial intelligence.
The study analyzed sequences from model organisms (yeast, mouse, and fruit fly) through deep learning and was able to determine and classify in great detail the functions of proteins that previously we had no information on.
Through deep learning, research groups have been able to determine the function of proteins for which only the DNA sequence was available.
The authors also found that, among the two deep learning methods used, language models or transformers are more efficient than convolutional networks. The latter are based on image processing, while transformers process sequences and language. This makes transfoer models more informative and precise, as well as able to retrieve information from RNA sequences.
This research is vital to addressing the issue of the dark proteome, which includes all proteins for which no information is available. In this way, proteins can be analyzed, and functions of genes with biomedical and biotechnological potential, especially in little-studied organisms, can be identified, says Rosa Fernández, co-leader of the study at IBE (CSIC-UPF). This is especially relevant now that unknown organisms are being sequenced in large quantities, leading to millions of sequences whose function cannot be predicted using traditional methods.
Israel Barrios-Núñez, Gemma I Martínez-Redondo, Patricia Medina-Burgos, Ildefonso Cases, Rosa Fernández, Ana M Rojas, Decoding functional proteome information in model organisms using protein language models, NAR Genomics and Bioinformatics, Volume 6, Issue 3, September 2024, lqae078, https://doi.org/10.1093/nargab/lqae078