When prescribing medication, a patient’s genetic information is often not considered. However, a deficiency in the G6PD gene, for example, can cause fatigue or abdominal pain when taking something as simple as an aspirin.
This is where pharmacogenomics—the study of how drugs affect people based on their genetics—becomes an emerging field. It may pave the way for personalized medicine where, in the future, simple genetic tests combined with artificial intelligence (AI) could identify vulnerable patients, or our pharmacogenetic profile could be included in our medical records.
Now, an international study led by the Institute for Evolutionary Biology (IBE: CSIC-UPF) has conducted one of the most extensive investigations into how genetic variation affects the toxicity of certain medications. This study used public databases, including the 1000 Genomes Project (1000G), the Simons Genome Diversity Project (SGDP), and the Human Genome Diversity Project (HGDP), to analyze genetic data from 3,714 people of various ancestries across all five continents. It focused on 1,136 genetic variants associated with drug toxicity. By using AI techniques, the team has been able to establish toxicity risk patterns that could be very useful in advancing personalized medicine.
“The fact that we observe differences between populations for these variants suggests it would be useful to consider an individual’s genetic ancestry when studying toxicity, to develop more personalized treatments.”
Òscar Lao, principal researcher of the study
The results reveal that populations of American and European ancestry have a higher risk of experiencing toxicity with a wide range of drugs. In particular, a greater susceptibility to cardiovascular and antimicrobial drugs has been observed in these populations. Furthermore, American populations show a high risk with antidepressants and painkillers, while Europeans are more vulnerable to immunosuppressants and anticancer drugs. On the other end, populations of East Asian ancestry and, to a lesser extent, Oceanian ancestry, generally show a low toxicity risk, except for some Central Asian populations that exhibit sensitivity to painkillers. African populations fall into an intermediate category, with a more diverse response.
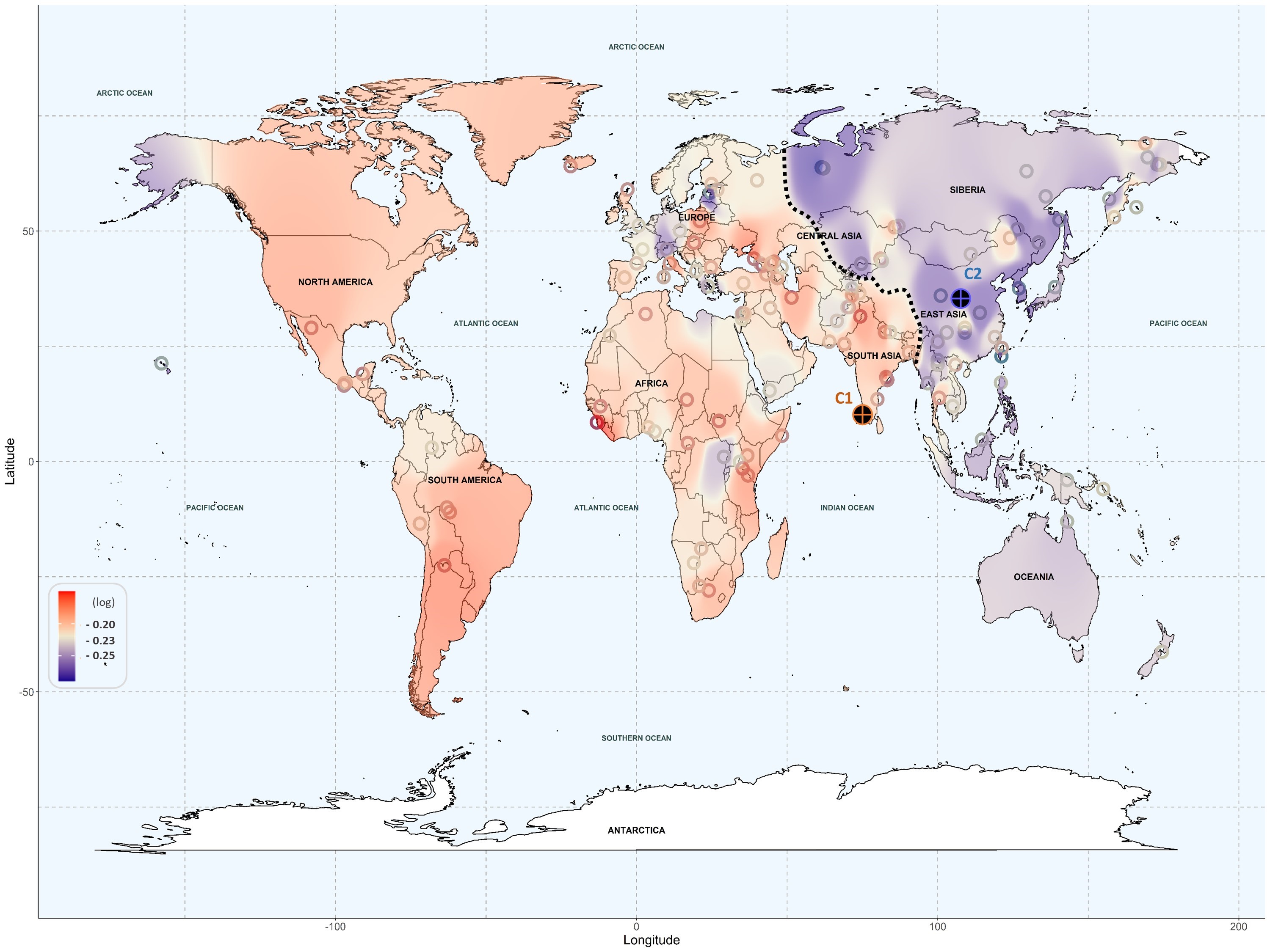
For the genomic analysis, researchers used machine learning techniques to study the function of genes involved in drug absorption and transport. Additionally, text mining—a technique for extracting and aggregating information from text data—enabled the grouping of genetic variants and their relation to specific genetic ancestries from certain geographical regions. These techniques have revealed genetic risk patterns that may significantly impact personalized medicine.
“Ultimately, however, the risk of adverse effects is specific to each individual, beyond their ancestry. Ideally, we should have our entire genome sequenced to establish our own risk patterns,” cautions Òscar Lao, principal investigator of the study.
Karamperis, K., Katz, S., Melograna, F., Ganau, F. P., Van Steen, K., Patrinos, G. P., Lao, O. (2024). Genetic ancestry in population pharmacogenomics unravels distinct geographical patterns related to drug toxicity, iScience, 27(10). DOI:10.1016/j.isci.2024.110916