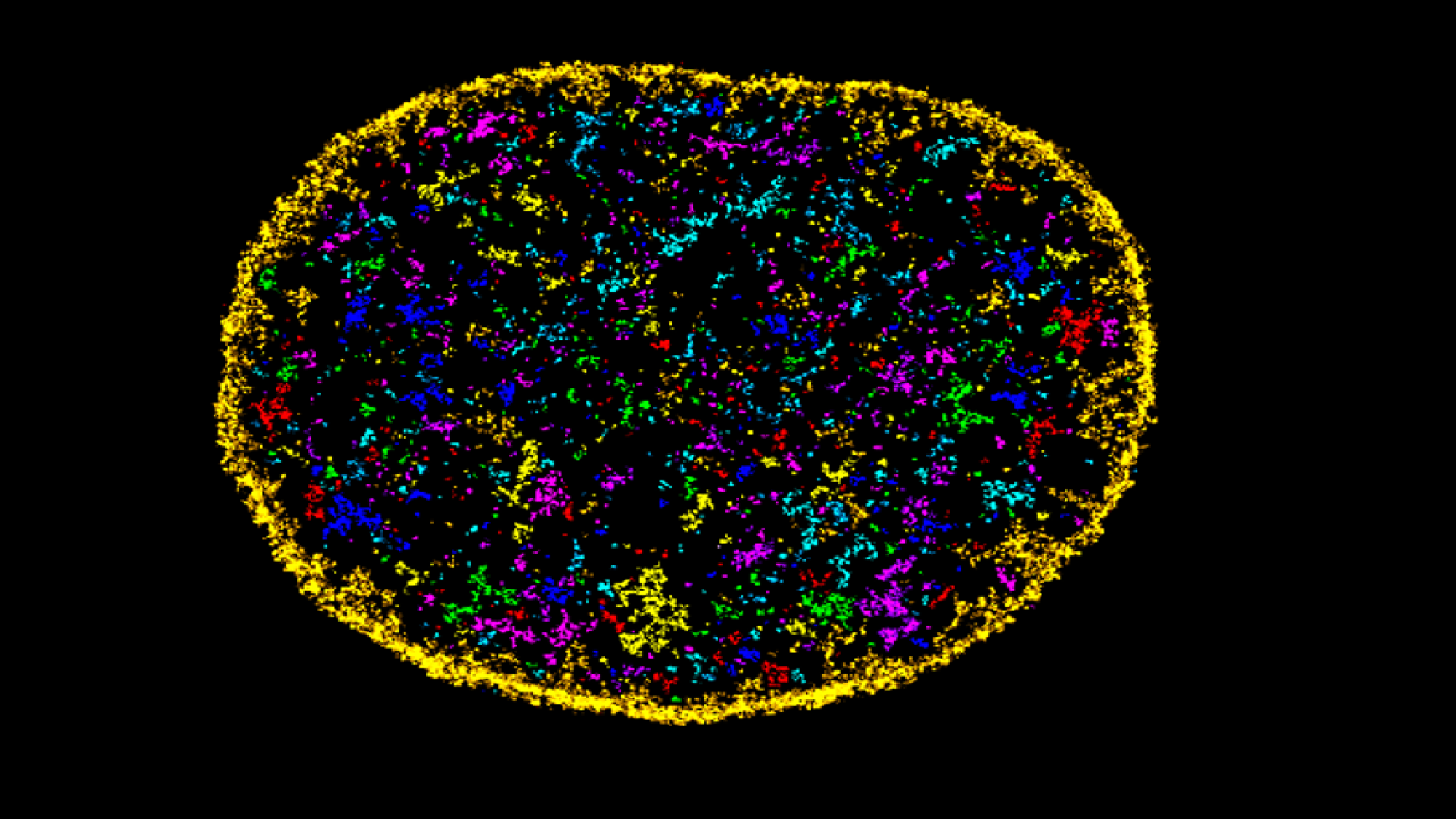
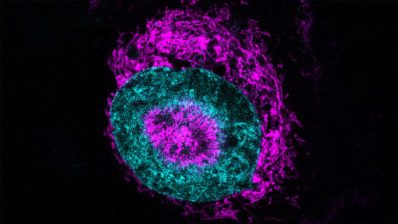
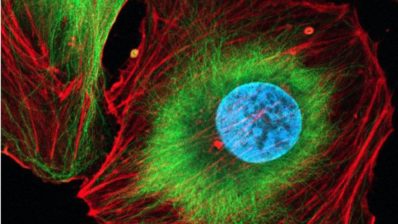
Researchers at the Centre for Genomic Regulation (CRG) and the University of the Basque Country have developed an AI tool, AINU (AI of the NUcleus), able to scan high-resolution images of cells, at nanoscale resolution and detect rearrangements as small as 20nm – that’s 5,000 times smaller than the width of a human hair.
“These rearrangements can be changes in how DNA is organised inside cells, or the distribution of enzymes within the nucleus, which both can change in cancer cells. Or they could be slight differences in how tightly DNA is packed after a virus infection”, says Pia Cosma, ICREA Research Professor and the CRG group leader behind the study.
Indeed, the AI was able to detect changes in a cell’s nucleus as soon as one hour after it was infected by the herpes simplex virus type-1.
The clinical applications of this tool are still probably years away, because it needs images taken with a special microscopy technique called STORM, which can only analyse a few cells at a time, and it’s a very specialized equipment which would require big investments.
But the researchers expect AINU to be already useful in scientific research, for example by identifying stem cells with very high precision, helping make stem cell therapies safer and more effective.
A detective algorithm to select the best drug
Another tool, RTDetective, has been co-developed between the CRG and the IRB. RTDetective is able to predict which drugs will be most effective in treating diseases caused by ‘nonsense mutations’, a type of mutations that act like a stop sign that prevents the synthesis of the full-length protein.
There are already “nonsense suppression therapies”, drugs which help cells ignore these stop signs and continue protein production. But not all work equally well for all mutations, because their ability to let the cell machinery ‘readthrough’ the nonsense mutation depends not just on the mutation itself, but also on the genetic code immediately surrounding it.
“A patient is diagnosed with a genetic disorder, the mutation identified through genetic testing and the best drug suggested by a computer model. This informed decision-making is the promise of personalised medicine we hope to unlock in the future”
Ben Lehner, ICREA Research Professor, CRG and Wellcome Sanger Institute
The researchers looked at 5,800 disease-causing premature stop signs and tested the effectiveness of eight different drugs on each of them. The data (140,000 individual measurements) was large enough to train accurate predictive models, which they used to create RTDetective.
The researchers next plan on confirming the functionality of proteins produced via readthrough drugs, a crucial step in validating their clinical applicability.
Carnevali, D., Zhong, L., González-Almela, E. et al. A deep learning method that identifies cellular heterogeneity using nanoscale nuclear features. Nat Mach Intell (2024). https://doi.org/10.1038/s42256-024-00883-x
Toledano, I., Supek, F. & Lehner, B. Genome-scale quantification and prediction of pathogenic stop codon readthrough by small molecules. Nat Genet (2024). https://doi.org/10.1038/s41588-024-01878-5