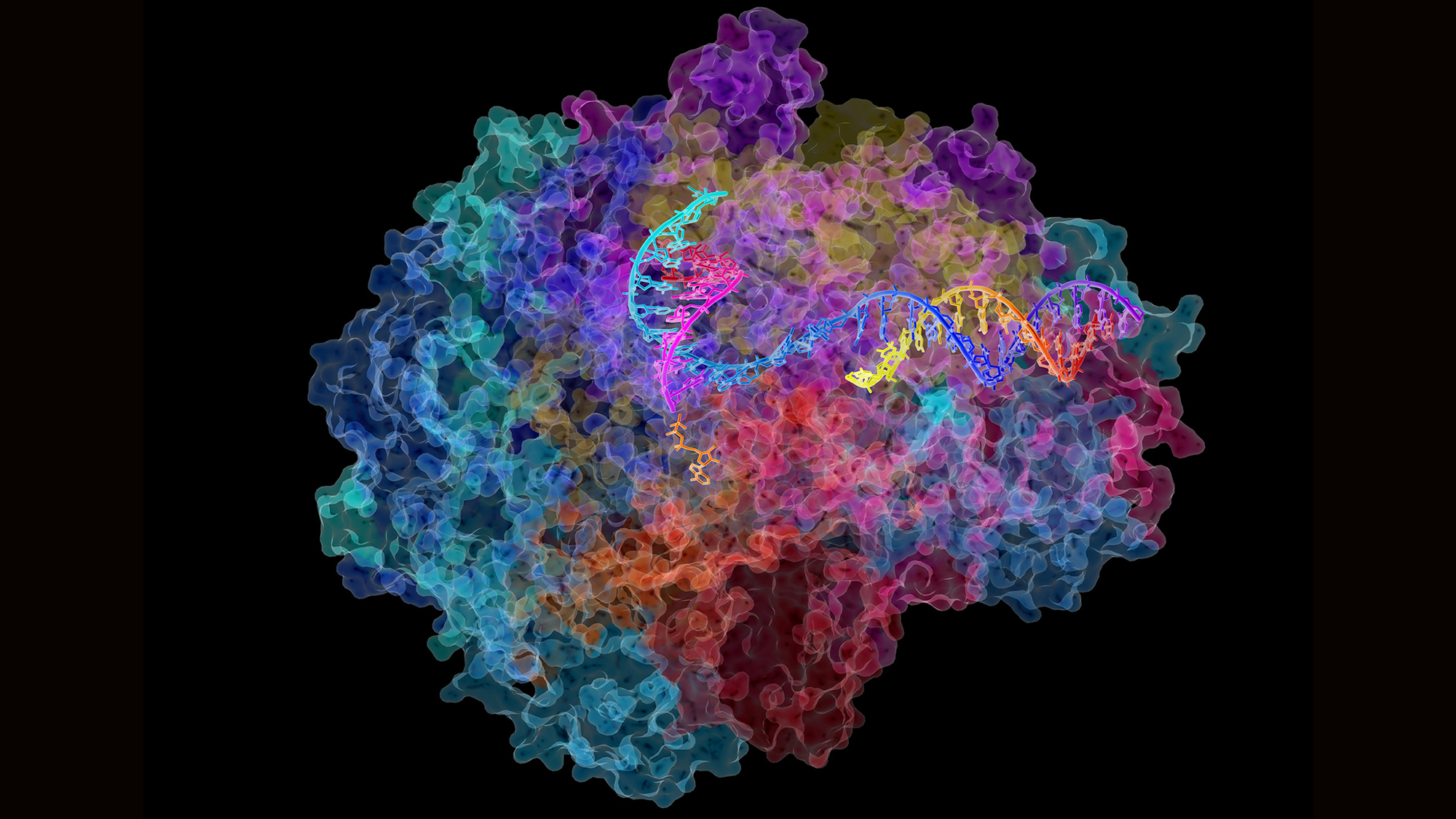
DNA can be defined as the instruction manual for life. It specifies the commands for making proteins, the ‘building blocks’ of living beings. When we say that a gene is expressed, it means a piece of DNA is ‘translated’ into a molecule of RNA, in a process called ‘translation‘, which is in turn transcribed into a protein, in a process called ‘transcription‘. For decades this was the dogma of molecular biology: each gene codes for a protein.
Tha basic dogma of molecular biology says that DNA is translated into RNA, which is then transcribed into proteins.
Some years ago, however, researchers discovered the existence of alternative splicing, a process where chunks of RNA are ‘cut and paste’ in different ways, meaning a single RNA can give rise to various proteins.
More recently, a further phenomenon has altered the canon once again. We now know that there are thousands of fragments of RNA that do not give rise to proteins. These are the so-called ‘non-coding RNAs’ (ncRNA), and there are many types. Transfer (tRNA) and ribosomal (rRNA) RNA were discovered between the 1960s and 1980s. But it is the last decade that has seen an exponential increase in these ncRNA, which have been categorised into different types according to their characteristics: microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNA, and so on.
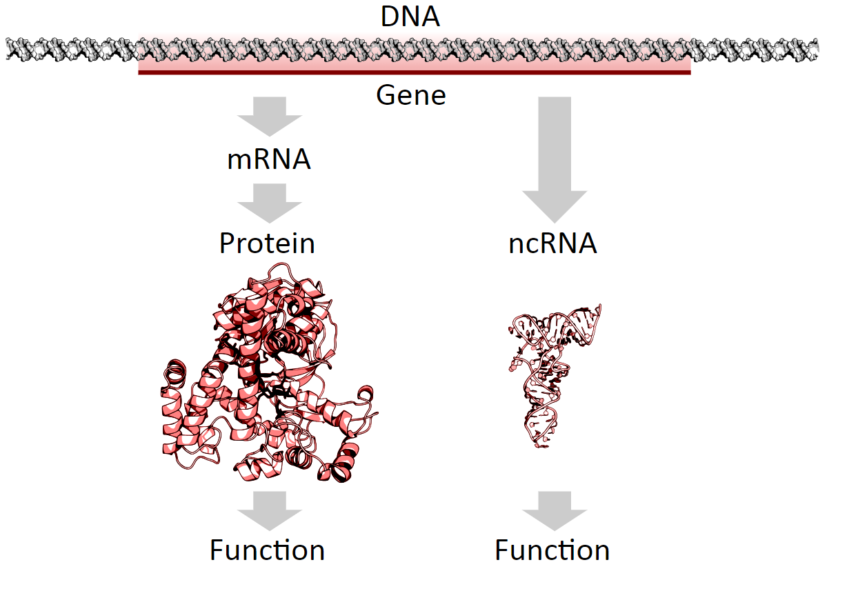
We now know that there are thousands of fragments of RNA that do not give rise to proteins. These are called ‘non-coding RNAs’ (ncRNA).
Although the functions of these ncRNAs are still not clear –some may not even have a function, and could simply be the result of a random translation of DNA– many of them are conserved in all species and are implicated in many important cell processes, including the regulation of gene expression, as well as in various diseases, like cancer, autism, and Alzheimer’s.