After 10 years of work, researchers from the Centre for Genomic Regulation (CRG) in Barcelona, led by Juan Valcárcel, have completed a groundbreaking study revealing the first blueprint of the human spliceosome, a crucial and complex molecular machine responsible for editing RNA in all human cells.
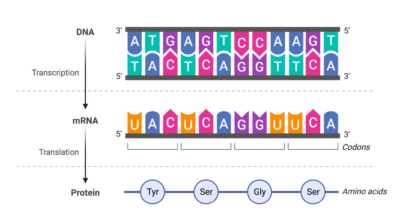
The spliceosome, composed of 150 proteins and five RNA molecules, plays a pivotal role in splicing, a process that allows cells to produce a diverse array of proteins from a single gene. Actually, more than 90% of the genes are subject to splicing, and splicing errors are linked to numerous diseases, including cancer and neurodegenerative conditions. But the intricate nature of the spliceosome has made it difficult to study until now.
The team at the CRG altered the expression of 305 spliceosome-related genes in human cancer cells one by one, observing the effects on splicing across the entire genome. This way they uncovered previously unknown specializations of spliceosome components, revealing their distinct roles in the splicing process.
Altering the expression of 305 spliceosome-related genes in human cancer cells, one by one, showed that individual components of the spliceosome are far more specialised than previously thought.
This new understanding could pave the way for targeted therapies, more effective and with fewer side effects, as many components that were once overlooked due to their unknown functions could now be considered for drug development.
“If we imagine the splicing process as the post-production of a film, the spliceosome is the studio where many dozens of editors are going through the material and making rapid decisions on whether a scene makes the final cut”, says Malgorzata Rogalska, co-corresponding author of the study. But there’s an unexpected twist in this ‘Hollywood production’. “Any of the contributors can take charge and become the director. And rather than the production falling apart, this results in a different version of the movie. It’s a surprising level of democratization we didn’t foresee”, she adds.
Another key discovery is that due to the high level of interconnectivity, disrupting any part of the spliceosome network can have cascading effects throughout the system. This could be leveraged to target cancer cells, where splicing machinery often becomes dysregulated. For example, alterations to the SF3B1 spliceosome component – which is mutated in many cancers including melanoma, leukaemia and breast cancer – led to widespread splicing failures in the cell, overriding its ability to continue growing. This could offer a potential “Achilles’ heel” for cancer treatment.
“One way cancers adapt is by rewiring their splicing machinery. If we obstruct this splicing, we could lead diseased cells past a tipping point that cannot be compensated for, leading to their self-destruction”
Juan Valcárcel (CRG), lead author of the study
The blueprint also promises to enhance our ability to diagnose and treat diseases caused by splicing errors. Currently, treatments targeting splicing defects are limited to rare diseases, but the new map could help researchers pinpoint exactly where the splicing errors are occurring and help extend these therapies to a broader range of conditions.
The CRG has made the blueprint publicly available, hoping to accelerate the development of new splicing-based treatments and make them more accessible to the mainstream medical community.
The study was done in collaboration with Remix Therapeutics, a clinical stage biotechnology company in Massachusetts.
Malgorzata E. Rogalska et al. Transcriptome-wide splicing network reveals specialized regulatory functions of the core spliceosome. Science 386,551-560(2024).DOI:10.1126/science.adn8105