Researchers from the Universitat Pompeu Fabra, Hospital del Mar Medical Research Institute (IMIM) and Universitat de Vic have developed a website to study the molecular environment of the proteins involved in diseases. The website is a promising tool to study the molecular mechanisms underlying a disease, to understand the relationships between two diseases and to propose potential drugs for their treatment.
Quim Aguirre, the PhD student behind this project, tells us all about it.
A network of interacting proteins
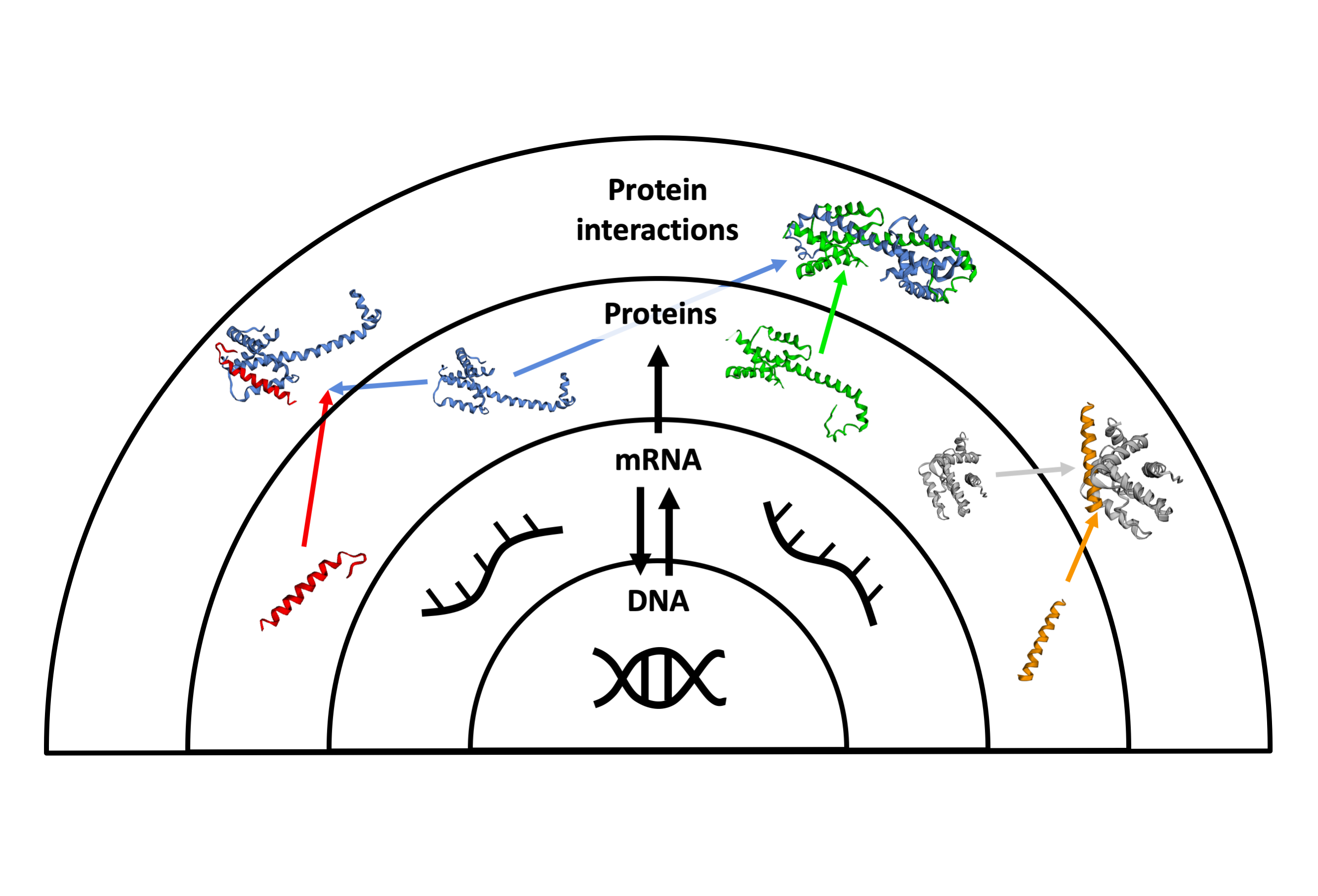
The human genome is formed by molecules that define the characteristics of each human being: the genes. Genes encode for proteins, the molecules that execute the specific functions to maintain the correct functioning of our body. Proteins interact with each other and with other biomolecules to successfully complete their functions. At a global view, each cell in the human body can be seen as a complex network of proteins specifically interacting with other proteins to execute the functions of the cell. This is often referred as protein interaction network or more recently as the human interactome.
Diseases: a tale of wrong interactions
However, the functioning of the proteins is not always correct. The alteration of genes, also called mutations, in some cases leads to an unusual behavior of the proteins and gives rise to diseases. Moreover, complex diseases such as cancer, diabetes, neurodegenerative disorders or cardiovascular diseases involve multiple mutations, and this complicates the study of their molecular mechanisms. Thus, the study of the protein interaction network is relevant to fully understand the molecular mechanisms that cause the disease and find potential treatments.
In search of the interactions underlying diseases
The website GUILDify has been developed at the Structural Bioinformatics Group (SBI) led by Prof. Baldo Oliva with the objective to study the protein interactions involved in a disease. When accessing the website, the user finds an input box to introduce any disease name and the species of the interactome (human, mouse, rat…). After introducing the disease, GUILDify provides the user with a list of proteins whose corresponding genes are likely to be mutated when the disease occurs. The gene mutations are obtained from DisGeNET, a database of genes associated to human diseases. GUILDify uses these proteins as seeds for an algorithm that searches for the proteins in the interactome that are more connected to them. In the end, the user obtains the protein interactions that are more likely to cause the disease and a list of the biological functions affected.
GUILDify uses the proteins known to be mutated in a disease as seeds for an algorithm that searches for other proteins in the interactome that are connected to them
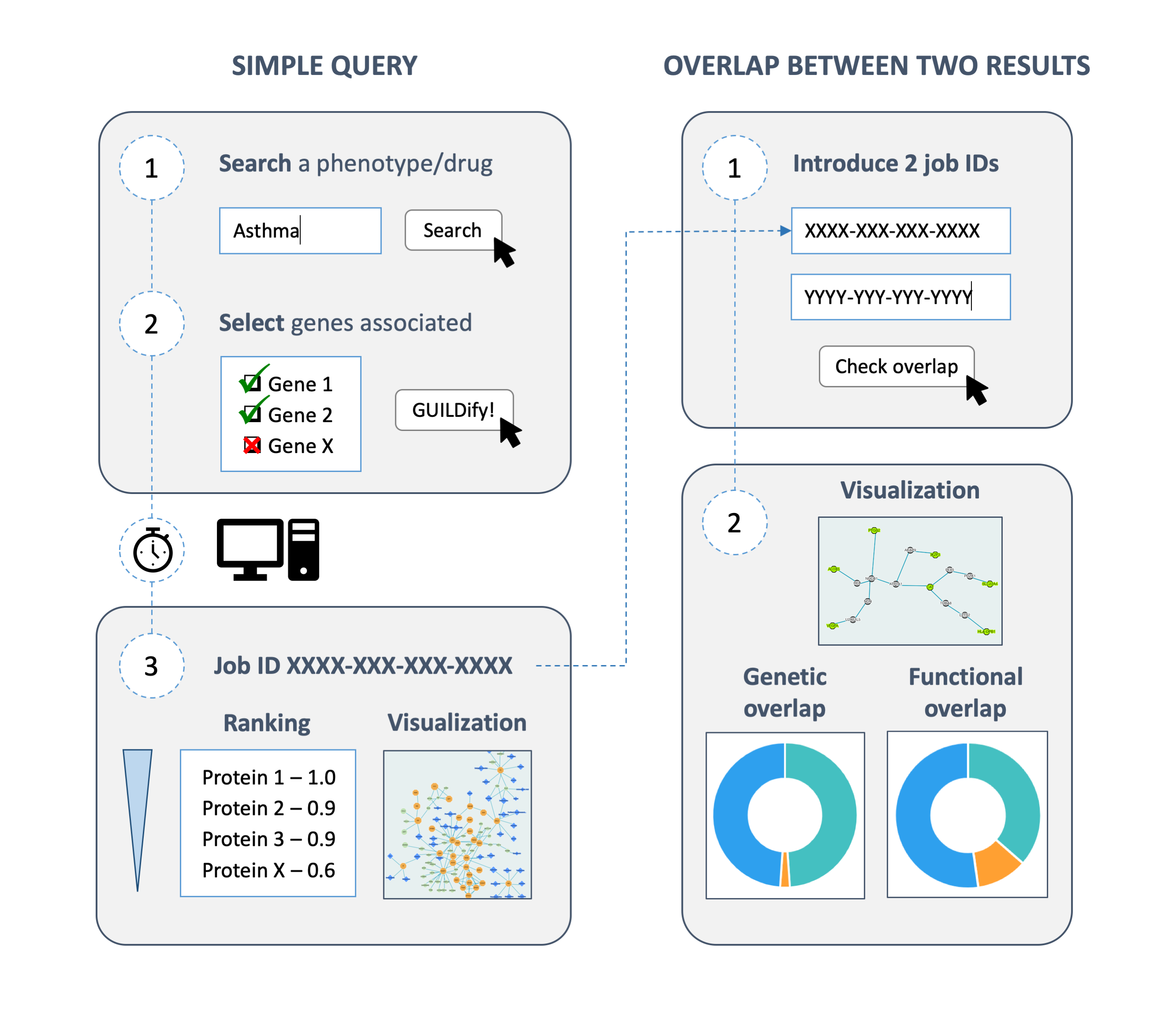
Apart from the study of diseases individually, GUILDify has also been adapted to study the relationship between two diseases. There are pairs of diseases referred to as comorbidities that are more likely to occur simultaneously in the same patient. In some cases, comorbidities can be explained because they share a common molecular mechanism. In a previous study by Carlota Rubio-Perez and colleagues at Department of Experimental and Health Sciences, Pompeu Fabra University (DCEXS-UPF) and IMIM, the software behind GUILDify was used to predict comorbidities successfully. Motivated by this research, the GUILDify website includes the option to introduce two diseases and explore the common proteins and biological functions involved.
From disease to treatment
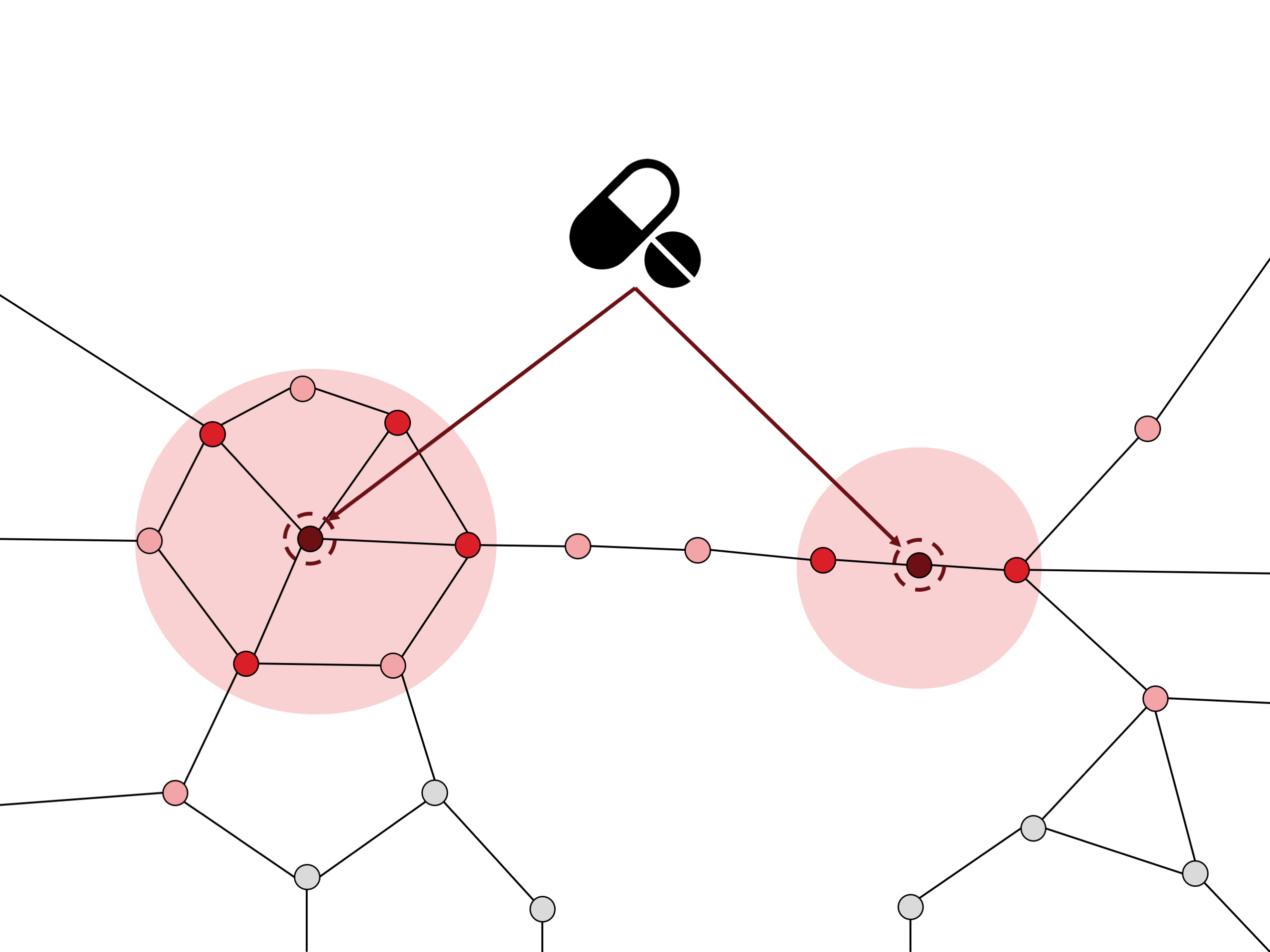
GUILDify also provides the option to study the molecular mechanisms of drugs. By providing a drug name to the search box, GUILDify provides the user with a list of target proteins which the drug interacts with to produce a therapeutic effect. In the same way as the diseases, it searches the proteins in the interactome that are more connected to the target proteins of the drug. The proteins found are the ones receiving a higher impact from the drug. Therefore, GUILDify can be used to see if the list of proteins affected by a disease is similar to the list of proteins affected by the action of a drug. This is an indicator of the impact that the drug has in the molecular environment of the disease.