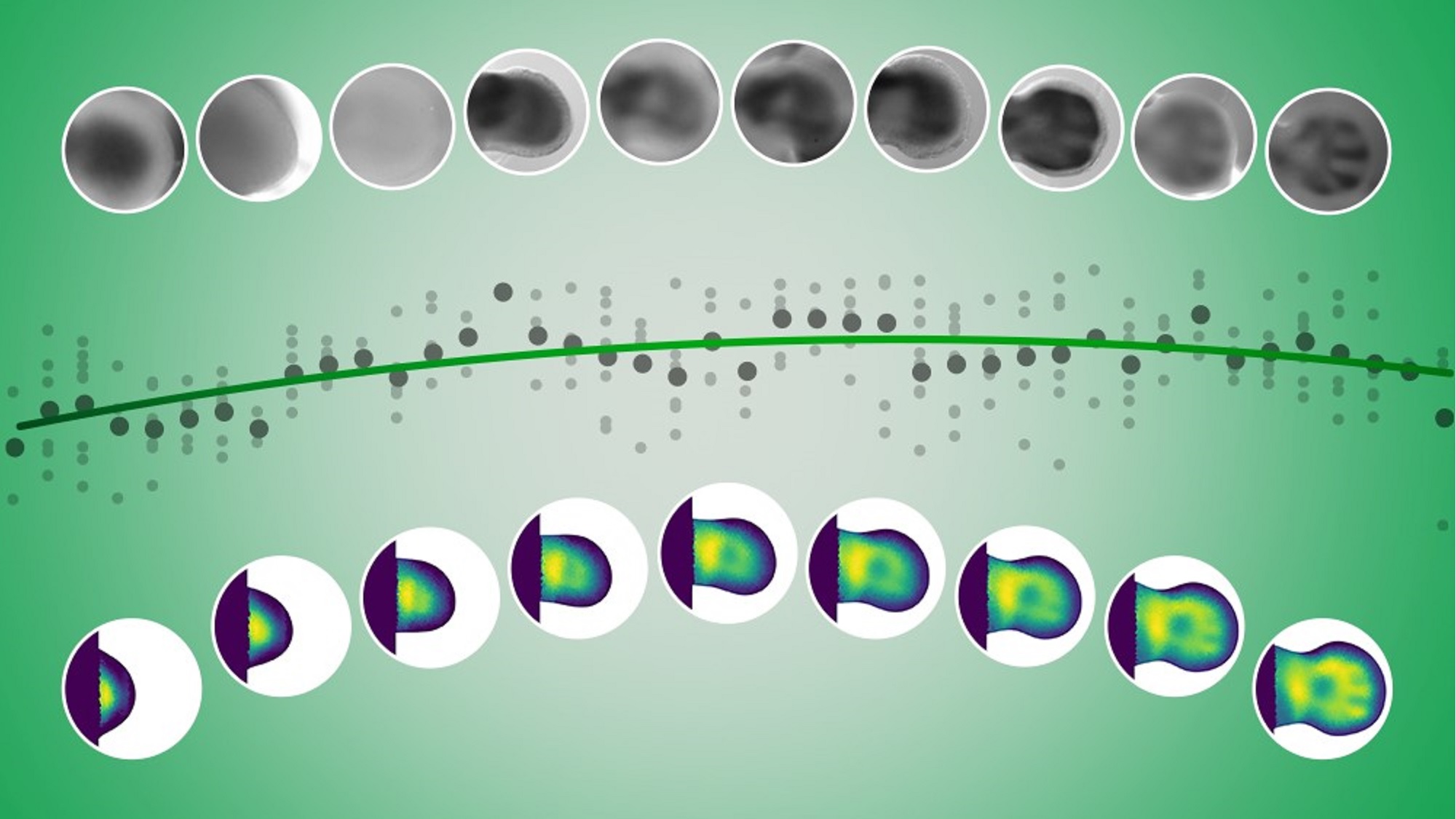
One of the major challenges in developmental biology is the difficulty of observing embryonic development in real time, especially in mammals, where organ formation occurs inside the uterus. Therefore, embryos must be studied through static snapshots at different stages, which provides a fragmented vision and requires large sample sizes.
The challenge becomes even greater when studying gene expression patterns, which are not tied to fixed anatomical structures, further complicating data interpretation.
To overcome these limitations, researchers in the Sharpe Group at the European Molecular Biology Laboratory – Barcelona (EMBL Barcelona) developed a new method to reconstruct a continuous timeline of gene expression patterns in mouse limb development.
Their approach focused on estimating the missing gene expression data points in the gaps between measurements. They did this interpolation for individual tissue segments before assembling them into a complete representation. This allowed them to capture the full dynamics of gene expression during development.
This method allows researchers to visualize gene expression changes dynamically without requiring additional data collection.
A key breakthrough came from an unexpected source—video games.
While she was playing a video game, PhD student Laura Aviñó-Esteban zoomed in into the smooth yet flexible interpolations game developers used for camera movements and animations. She then did some research that led to the team adopting the mathematical functions commonly used in computer graphics (called B-Splines) to create the gene expression reconstructions.
Their method was successfully applied to Sox9, a gene involved in skeletal development, as well as other processes like neural tube formation, demonstrating its broad applicability.