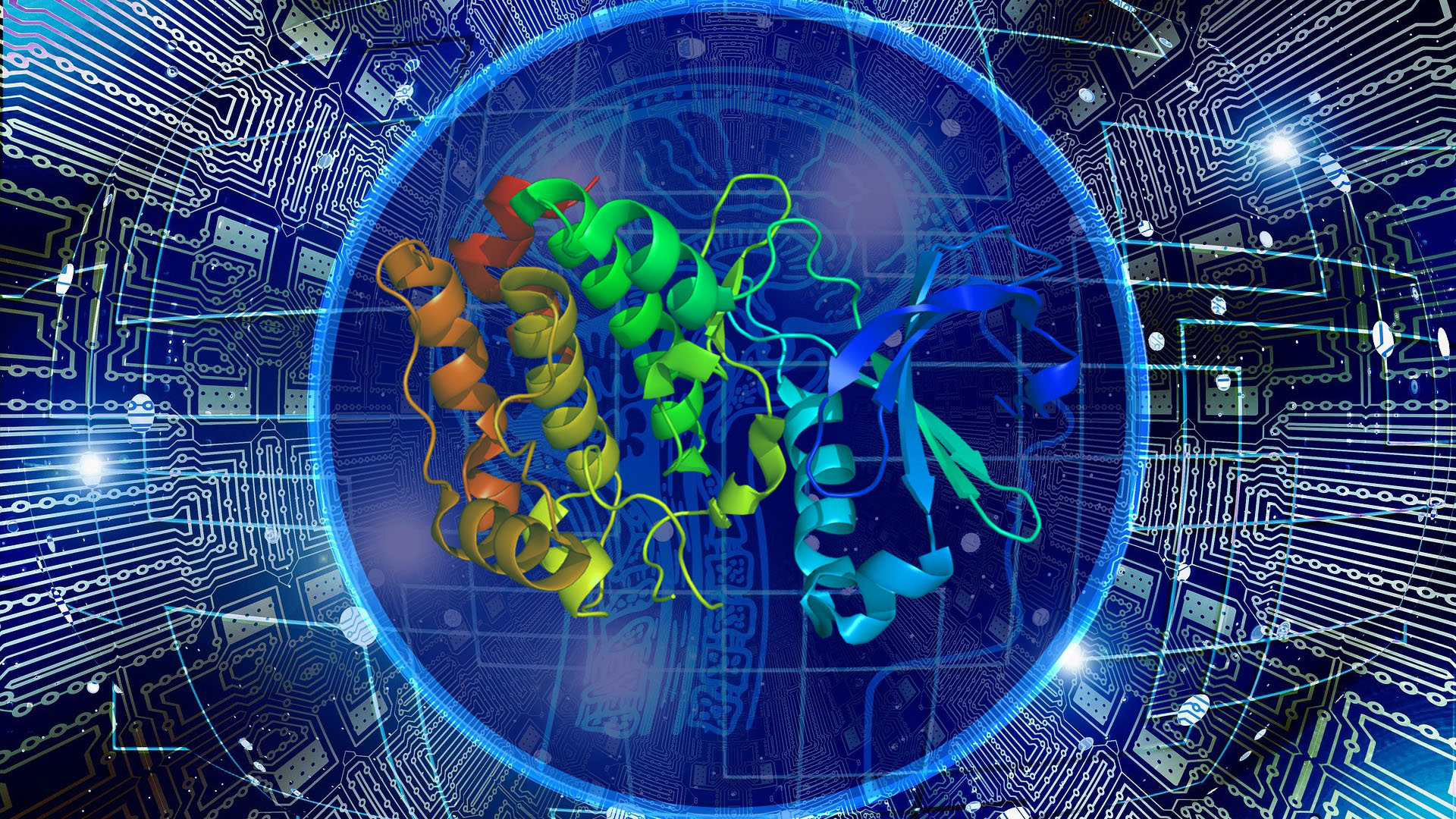
Knowing the structure of proteins – the building blocks that form cells – is essential to understand their function and, therefore, to understand how organisms work, and how and why diseases occur.
There are two main ways to find the structure of a protein:
- Experimental: through cristallography, MR, etc.
- Theoretical: with computers, comparing the sequence to those of proteins with known strcucture, etc.
For more than 25 years, the international biannual CASP (Critical Assessment of protein Structure Prediction) competition has joined experimentalists and theoretical researchers to test the predictive models of the latter.
Only a few years ago, hitting 10% of the proposed protein structures was considered a success, and two years ago the Google team surprised the scientific community by hitting 50%. In the most recent CASP competition, the result of which was announced at the end of 2020, the company DeepMind– belonging to Google – used its AlphaFold artificial intelligence system and achieved a correct prediction of 100%, to the great surprise of all participants, causing a revolution in this field.
In the latest episode of the podcast aimed at the general public, “Peter Parking”, Baldo Oliva, leader of the structural bioinformatics group of the GRIB, at the Barcelona Biomedical Research Park (PRBB), explains how artificial intelligence (and specifically deep learning) has just transformed the prediction of protein structure.
You can listen to the podcast in Spanish here: