Today, 18 years after having sequenced the first human genome, what can our genome really tell us? A lot, according to two outstanding scientists working at the Barcelona Biomedical Research Park (PRBB): Jaume Bertranpetit from the Institute for Evolutionary Biology (IBE: CSIC-UPF) and Roderic Guigó from the Centre for Genomic Regulation (CRG). Both shared the stage at the PRBB auditorium in a session that had been organized by Juan Valcárcel from the CRG.

The main question that captures the interest of Jaume Bertranpetit, head of the Evolutionary Systems Biology lab, is how to reconstruct the past in a system that evolves. His research group now uses deep learning algorithms to understand the potential influence of different demographic events on our genome.
One of the most recent results of this approach is the identification of the genetic footprint of a new hominid in Asian populations who cross bred with our ancestors tens of thousands of years ago. Bertranpetit suggests that this extinct hominid was a hybrid of Neanderthals and Denisovans and cross bred with “Out of Africa” modern humans in Asia.
Other readings from our genomes appear to be much more certain, like the fact of a single origin of the out of Africa migrations of the humans, or the finding that we all have a piece of Neanderthal genome, except for the Africans.
Positive selection can be detected in the genome
In the second part of his talk, Bertranpetit explained his most recent work on positive selection and adaptation. Genome-wide association studies (GWAS) are providing a lot of data since they measure genetic variants in different individuals to see if any variant is associated with a trait. Most of these data are accessible in the 1000 Genome Browser, the largest public catalogue of human variation and genotype data.
Ongoing projects include the appearance of milk intolerance, positive selection of a taste receptor that tastes sour in African populations and traits like skin colour and height. The genes responsible for the height of a person are known now, which led to a finding suggesting that dwarfism is a positive selection strategy for island populations.
As a conclusion, Bertranpetit’s talk confirms that the DNA harbours information about our history and claims that positive selection can be detected in the genome, although it is difficult to interpret the potential findings.
Next it was the turn of Roderic Guigó, coordinator of the Bioinformatics and Genomics programme at the CRG. He was introduced by his longstanding collegue Jaume Bertranpetit, saying that he certainly had demonstrated that one can learn a lot of biology by just reading the genome.

The path from DNA to organism is very complex and according to Guigó not as easy as it is illustrated in an adaptation of “La Clairvoyance” from René Magritte that Bertranpetit used as a slide in his early teachings. In spite of all the GWAS data and more than 25,000 SNP traits, scientists know little about how an individual phenotype is formed. To be able to visualize an organism with all its observable characteristics (phenotype) just by looking at the DNA sequence, as suggested in the painting, is still a fantasy.
To be able to visualize an organism with all its observable characteristics (phenotype) just by looking at the DNA sequence is still a fantasy.
RNA is the first phenotype and all the sets of RNA in a cell, tissue or organism, the transcriptome, have been studied by the GTEx Consortium with the participation of Guigó’s research group. The scientists have analysed the transcriptome of 17,382 samples, from 55 different tissues and 948 individuals.
If one looks at sex biased gene expression, there are some surprises. Hardly any difference was found in the analysed female and male tissues. Not even in breasts. Males just lack lobules to produce milk.
Another finding was that there is an unexpected diversity of expression patterns in all tissues. The lung, for example, shows the same internal expression diversity than other organs of the human body. The reason is that each organ is composed of many different types of cells. In order to bring more light into this matter, scientists are now proceeding with single cell analysis of expression for the different tissues.
One of the remaining questions is how cells can actually form organs and what the mechanisms to enable this process are. For a better understanding of the diversity of human tissues, Guigó recommends exploring the GTEx catalogue of histology images in high resolution of all tissues.
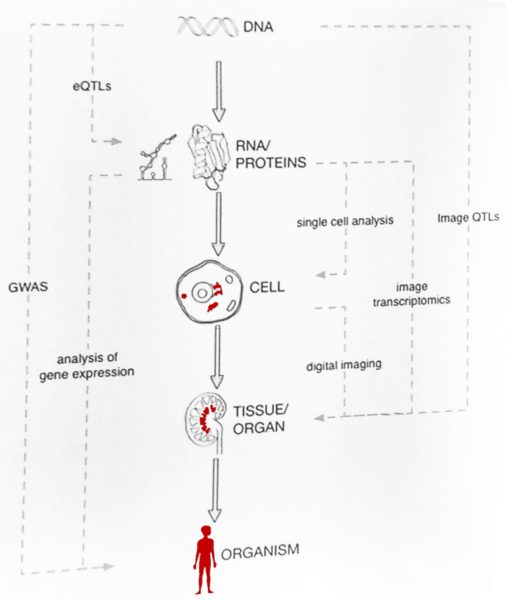
Guigó finished his talk with a few more milestones in the understanding of the long path from DNA to organism. Considerable progress has been made not only with single cell analysis (see the Human Cell Atlas), but also with digital imaging of tissues and organs from healthy and sick donors. This method can also identify the “transcriptional” cell types: epithelial, endothelial, blood, mesenchymal and neuronal, within the tissue samples of blood vessels, kidneys or tumours. Connecting the data from genotype, phenotype and histology promises exciting new results in the near future.
Mayukh Mondal, Jaume Bertranpetit, Oscar Lao. Approximate Bayesian computation with Deep Learning supports a third archaic introgression in Asia and Oceania; Nature Communications 10, Article number: 246 (2019) DOI: 10.1038/s41467-018-08089-7
GTEx Consortium, et.al. Genetic effects on gene expression across human tissues. Nature. 2017 Oct 11; 550(7675): 204-213. doi: 10.1038/nature24277